At present, the molecular profile of solid tumours is established using surgical or biopsy tissue samples. Tissue-based tumour profiles are, however, subject to sampling bias, provide only a snapshot of tumour heterogeneity, and cannot be obtained repeatedly. Analyses of circulating nucleic acids, commonly referred to as 'liquid biopsies', can be used to monitor response to treatment, assess the emergence of drug resistance, and quantify minimal residual disease. In addition to blood, several other body fluids can contain tumour-derived genetic information. In an article published in the Nature Reviews Clinical Oncology the research experts examine how different forms of liquid biopsies could be integrated into clinical practice, focusing on liquid biopsy of cell-free tumour DNA (ctDNA), the most clinically advanced approach.
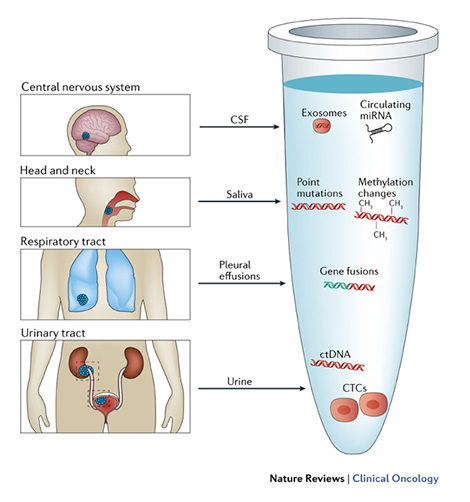
The molecular profile of tumours evolves over time. Human blood samples contain cell-free DNA (cfDNA) and RNA (cfRNA), proteins, cells, and vesicles (such as exosomes) that can originate from different tissues, including cancers. The rapid turnover of cancer cells is postulated to result in the constant release of tumour-derived nucleic acids and vesicles into the circulation, and viable tumour cells can also separate from the tumour to enter the bloodstream. Thus, the ability to detect and characterize ctDNA and/or tumour-derived RNA (predominantly microRNAs), and circulating tumour cells (CTCs), has enabled clinicians to repeatedly and non-invasively interrogate the dynamic evolution of human cancers.
In addition to blood, some other body fluids such as urine, saliva, pleural effusions, and cerebrospinal fluid, as well as stool, have been shown to contain tumour-derived genetic material, and our ability to exploit liquid biopsies for diagnostic purposes will further expand in the future. The analysis of liquid biopsy specimens is, however, challenging because ctDNA is fragmented and highly under-represented compared with germ line cfDNA, and only a limited number of CTCs can be isolated from a blood sample. Thus, analysis of tumour material obtained by liquid biopsies requires highly sensitive assays, which became available only recently.
In their article, the authors provide an overview of the various types of tumour-derived material that can be sampled using liquid biopsies. Subsequently, they discuss how different forms of liquid biopsy can be exploited in patient care. Applications of liquid biopsies in oncology have emerged and developed at an incredible rate over the past 5 years. Many clinical trials performed in patients with solid tumours now incorporate longitudinal blood collections; however, most of the studies have included only small cohorts of patients.
Implementation of liquid biopsy approaches in clinical practice will occur only after extensive controlled studies are performed. Currently, more than 60 trials, with a projected accrual of more than 20 000 patients across 11 cancer types, are addressing the challenges posed by liquid biopsy.
Exploiting liquid biopsy approaches in patient screening could provide a more comprehensive view of tumour characteristics, including aggressiveness and the overall molecular landscape. Comprehensive implementation in the clinical setting will probably include the initial analysis of a subset of candidate genes for known hotspot mutations using PCR-based approaches, as a benchmark test and to confirm the presence of sufficient amounts and quality of ctDNA, followed by broader ctDNA sequencing to uncover actionable therapeutic targets.
The authors conclude that the next generation of 'liquid biopsy' studies will be key to definitively establish the clinical applicability of blood-based genomic profiling. They wrote: “Liquid biopsy approaches will probably provide improved diagnostic power, but a key question remains: will liquid-biopsy-driven treatments translate into improved outcomes?”